A Harmonious Blend of Biology and Coding: Utilizing Python
Written on
Chapter 1: The Union of Biology and Programming
Have you ever pondered the fact that nearly 50% of marriages in the United States end in divorce? Thankfully, the partnership between biology and programming isn't one of those failed unions, and I aim to demonstrate this by the conclusion of this article.
Previously, I investigated DnaA boxes within the ori region of V. cholerae. However, what if the location of ori within the genome is unknown? The key lies in transforming biological knowledge into computational solutions. (This article assumes a solid understanding of Python and DNA replication.)
“Let’s discuss DNA replication, bay-bee!” — Salt-N-Pepa, 1990.
Bacteria replicate their genomes in an asymmetric manner. One half is continuously synthesized: DNA polymerase links nucleotides seamlessly. In contrast, the other half is produced in a discontinuous fashion: numerous short DNA segments are synthesized and subsequently joined to create a continuous strand.
Bacterial chromosomes are circular, and DNA replication initiates at the ori region. Thus, we can segment the chromosome into four half-strands:
- Reverse half-strands: These serve as templates for the continuous formation of forward half-strands, proceeding in a 3' to 5' direction from ori.
- Forward half-strands: These act as templates for the discontinuous synthesis of reverse half-strands, moving in a 5' to 3' direction from ori.
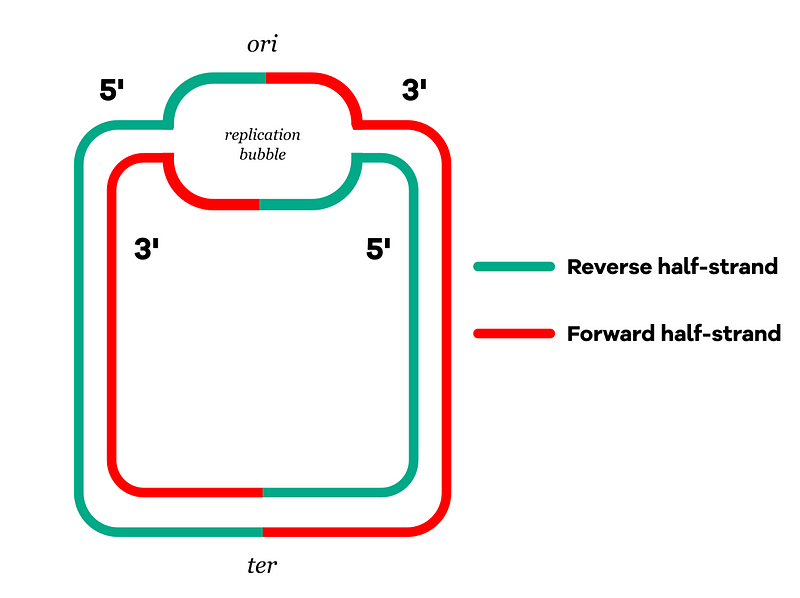
Replication occurs from both sides of the replication bubble and concludes at the ter region, situated opposite to the ori region. In the video, did you observe how the forward half-strands are frequently single-stranded? This increases the likelihood of cytosine deaminating (mutating) into thymine.
From this information, we can draw three conclusions:
- Deamination occurs repeatedly throughout evolutionary history, leading to forward half-strands typically having reduced cytosine.
- When reverse half-strands are synthesized on mutated forward half-strand templates, DNA polymerase tends to incorporate adenine (which pairs with thymine) instead of guanine (which pairs with cytosine). Consequently, reverse half-strands will often contain less guanine.
- Most crucially... In the 5' to 3' direction along the genome, the reverse half-strand converges with the forward half-strand at the ori region. “I’m askew da-ba-dee-da-ba-daa” — Eiffel 65, 1998.
Let’s denote the difference between the number of guanines and cytosines as 'skew':
skew = #G - #C
- On the forward half-strand, #G > #C
- On the reverse half-strand, #G < #C
By analyzing windows across the genome, if skew is increasing, it indicates a reduction in cytosine — we are on the forward half-strand. Conversely, if skew is decreasing, it signifies fewer guanines — we are on the reverse half-strand.
The Code
Since ori is located where the reverse half-strand meets the forward half-strand, we can determine ori by identifying the point where skew transitions from decreasing to increasing, also known as minimum skew.

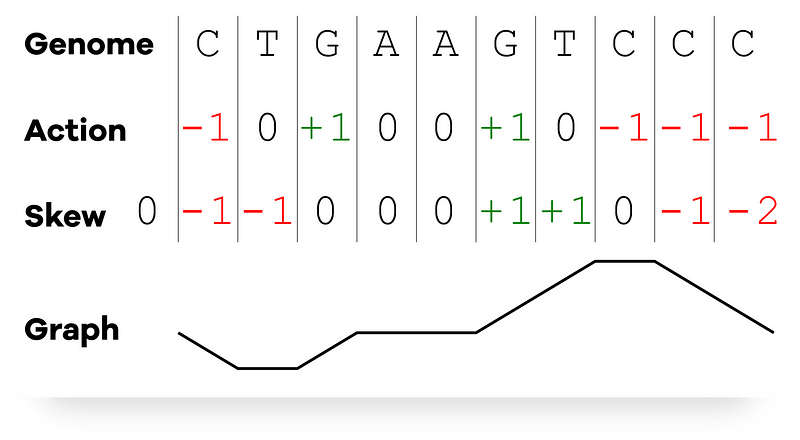
Begin by taking a string Genome as an argument. We'll create a list Skew where the initial value is 0. Each element in Skew will represent a running tally of #G — #C at each position in the genome. At the 0th nucleotide (prior to the first nucleotide), Skew = 0.
Next, we iterate through the genome, comparing each character in Genome to “G” or “C”:
- If it’s “G”, we increment the running count for skew and append it to the list Skew.
- If it’s “C”, we decrement the running count and do the same.
- If it’s neither, the skew remains unchanged.
Here’s a diagram illustrating this process:
An alternative implementation of this function (credit to Cristian Alejandro Fazio Belan) is more intuitive:

In this version, we update the cumulative score and add it to the list.
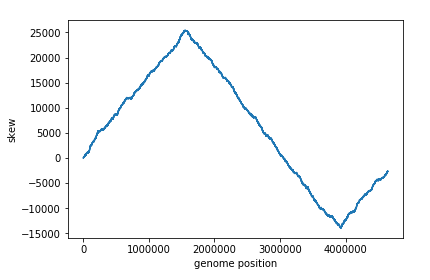
We can then plot this list to visualize the skew:
To find the point of minimum skew, we can write a quick function that returns all positions where Skew reaches its minimum:

>>> positions = MinimumSkew(Genome)
>>> print(positions)
[3923620, 3923621, 3923622, 3923623]
Alternatively, you can simply do the following:
>>> skew = SkewArray(Genome)
>>> print(skew.index(min(skew)))
3923620
For E. coli, it appears that ori is located around the 3923620th nucleotide. But how can we verify this?
In a previous article, we learned how to identify DnaA boxes within a genomic sequence. These are typically 9-base sequences (9-mers) that DnaA proteins bind to initiate DNA replication.
If we slice 500 bases from the point we’ve identified (the average length of ori) and apply the FrequencyMap and FrequentWords function from the earlier article…
>>> for i in range(3,10):
print(i, ":", FrequentWords(Text, i))
We may not find any 9-mers or reverse complements that appear three or more times.
(FrequencyMap scans a sequence and constructs a dictionary containing all possible k-mers as keys, with their frequencies as values. FrequentWords returns the most common k-mers.)
Once again, biological knowledge comes to our aid. DnaA proteins can bind to DnaA boxes that are approximately similar. Two 9-mers with slight mismatches can still be recognized by DnaA.
The number of mismatches can be referred to as the Hamming distance. Let's write a function to determine this:
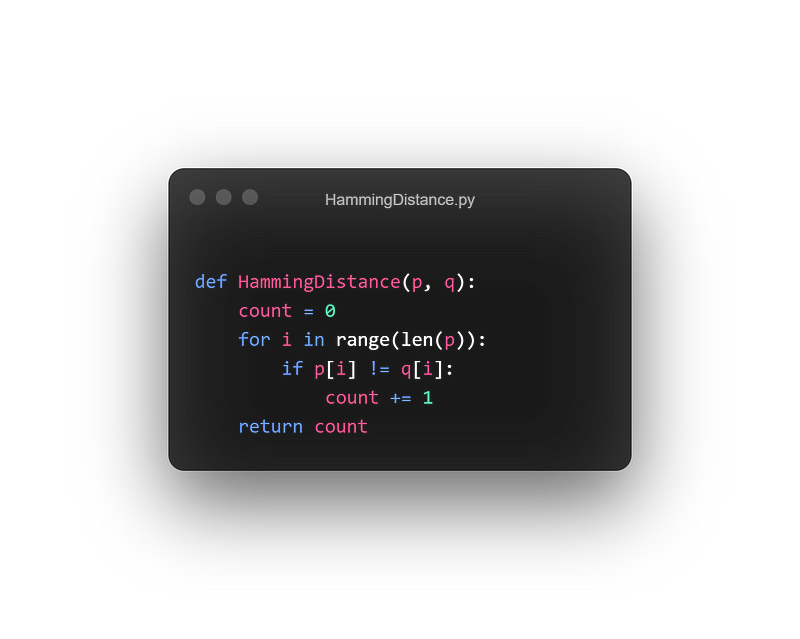
Take two strings p and q as parameters and iterate through each character. If they differ, increment the count.
We can utilize the 9-mers from our FrequencyMap as inputs in an updated PatternCount function:

Here, Pattern is one of our 9-mers, Text is the 500-base slice we took at ori, and d represents the maximum number of mismatches allowed.
We traverse the hypothetical ori region and compare a window against the Pattern, any of the identified 9-mers or their reverse complements. If they exhibit one or fewer mismatches, we increment the count.

This function may not be the most efficient, but it effectively accomplishes the task.
Ultimately, we discover TTATCCACA. Its reverse complement, TGTGGATAA, are the experimentally identified DnaA boxes for E. coli! However, they are not the only candidates. Other 9-mers appear with similar frequency. So, why is this significant?
By gathering several potential 9-mers and their genomic positions, we can provide valuable insights to researchers and save them time in the laboratory.
Programming helps clear the clutter, enabling biologists to locate the crucial information more efficiently. The partnership between biology and computer science is unique in that it frequently proves to be fruitful.
I got you there!
Comprehending biology allows us to develop effective and practical algorithms. Utilizing programming alongside biological data empowers biologists to enhance their experiments.
If you can navigate the intersection of biology and computer science, you possess extraordinary skills. You can build specialized expertise and likely achieve significant success. Most importantly, you can contribute to vital work that benefits humanity.
Thank you for reading! This article draws from a lesson in the Bioinformatics for Beginners course I’m currently taking on Coursera.
Chapter 2: Enhancing Understanding with Video Insights
The first video title is A Match Made in Heaven: Using Computation to Find Better Antibodies - Ariel Tennenhouse. This presentation explores how computational methods can enhance the search for effective antibodies.
The second video title is This DNA Discovery Is Completely Beyond Imagination | Gregg Braden. In this talk, Braden discusses groundbreaking revelations in DNA research that challenge conventional understanding.